
By: Amanda Chung
As the world began to lock down in the early months of 2020 at the onset of COVID-19, researchers in the McLellan Lab at The University of Texas at Austin were furiously at work.
Part of the Molecular Biosciences Department, the McLellan Lab is a team of graduate students and postdoctoral scholars led by Jason S. McLellan, Professor of Molecular Biosciences and the Robert A. Welch Chair in Chemistry at UT Austin. The lab examines the structural mechanisms of pathogens to develop therapeutic interventions that prevent disease caused by viruses, bacteria and other microorganisms.
McLellan Lab members worked nonstop to use the genetic sequence of the SARS-CoV-2 virus and unique skills and knowledge at UT Austin to develop a stabilized version of the spike protein, the feature of the COVID-19 virus that enables it to penetrate host cells and cause infection. Genetically altering the spike protein to stabilize it allows for its use in a vaccine that triggers a powerful immune response against the virus.
The hard work of the scientists in the lab paid off: Within just three weeks of obtaining the SARS-CoV-2 genome sequence, the team had successfully mapped the structure of the spike protein and created an antigen—the part of a vaccine that provokes an immune response and makes it effective.

Jason S. McLellan, professor of molecular biosciences and the Robert A. Welch Chair in Chemistry at UT Austin, poses in a lab on campus on June 30, 2021.
Today, hundreds of millions of people across the world have been immunized by vaccines that use the McLellan Lab’s technology. The contributions of the many talented graduate students in the lab were instrumental to this achievement. “Graduate students played a critical role in determining the first structure of the SARS-CoV-2 spike protein as well as the development of our second-generation stabilized spike protein, HexaPro,” McLellan says. “We had to move quickly given the urgency of the situation, and every day mattered. Many graduate students worked around the clock to clone, express, purify and characterize the spike proteins we had designed. This effort … would not have been possible without all of the dedicated graduate students who worked on this project.”
Springing into Action
Ph.D. students were among the first members at the McLellan Lab to immediately apply their research to help the fight against COVID-19. Daniel Wrapp—a Ph.D. student in cell and molecular biology at the time and current postdoctoral fellow at the Duke Human Vaccine Institute—set out to determine whether lessons learned from other coronaviruses could be used against the spike protein from COVID-19.
Fellow cell and molecular biology Ph.D. student Jory Goldsmith and Postdoctoral Fellow Nianshuang Wang assisted Wrapp and produced SARS-CoV-1 antibodies in the lab to chemically test their affinity for the SARS-CoV-2 spike. “We provided the first evidence that SARS-CoV-1 antibodies could not stick to the isolated spike protein of SARS-CoV-2,” Goldsmith says. “This meant that the new virus was different enough from the 2001 SARS virus that people with antibodies against SARS-CoV-1 would not be protected.”

Nianshuang Wang, research associate, right, and Daniel Wrapp, graduate student, left, review cryo-EM images in The Sauer Structural Biology Laboratory in February 2020.
In 2017, when the lab was then based at Dartmouth College, researchers used a two-proline mutation to stabilize the structure of the spike protein of the Middle East respiratory syndrome (MERS) coronavirus. This mutation locked the otherwise shape-shifting protein in place, allowing for the elicitation of more neutralizing antibodies. Solving and stabilizing the MERS spike protein had resulted in the lab’s establishment of the protocols, techniques and skills necessary to produce a specific spike-protein shape at the microscopic level.
These innovative methods and foundations helped Wrapp, Wang and their colleagues to quickly determine the SARS-CoV-2 spike structure about three weeks after receiving the genome sequence of the virus. “Both the knowledge of the stabilizing mutations and the fact that our lab had been producing MERS and SARS spikes for years allowed us to rapidly produce the SARS-CoV-2 spike and determine its shape,” Goldsmith says. “Producing a spike protein that actually has the correct shape at the microscopic level is not trivial, but since we had been optimizing our protocols, techniques and skills, that part was pretty routine. Daniel was prepared to solve the SARS-CoV-2 spike structure before he even knew about it.”
Refinement and Improvement
It was around this time that the lab began to develop HexaPro, an even more stable version of the spike protein. HexaPro utilizes six molecular mutations, rather than two, for a more durable antigen, making it a good candidate for use in parts of the world where the ultra-cold storage needed for mRNA vaccines can be a barrier.
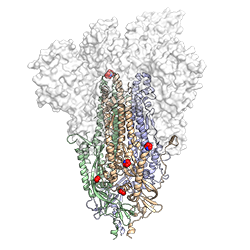
Identifying which specific mutations worked best out of the endless combinations possible required strategic organization for testing in the lab. Postdoctoral Fellow Ching-Lin Hsieh teamed up with McLellan to design 100 potential stabilizing mutations before delegating initial testing across a group of graduate students and postdoctoral fellows in three different UT labs. “While Dr. McLellan and lead author Ching-Lin designed most of the mutations that would potentially further stabilize the spike protein, other postdocs and graduate students helped to produce the newly designed constructs and test them for more robust expression,” says Nicole Johnson, a Ph.D. student in biochemistry.

Goldsmith worked in the lab alongside other graduate students to expedite the SARS-CoV-2 spike protein research process during the height of the pandemic. He supported the projects by performing biochemical and structural experiments to identify optimal stabilizing substitutions.
The incredible pace of work maintained by the students was the result of clever planning to maximize the collaborative effort of the lab. Each student performed their respective role as quickly as possible to relay information needed for the next step in development.
Johnson says this phase of development was one of the most challenging stages of the project. The overwhelming number of potential combinations that the lab had to analyze posed a serious threat to their swift process. Luckily, the lab’s pre-existing organization system and collaborative culture made for quick and easy distribution of work. “We had so many people involved that made it possible and pretty seamless to divide and conquer. We all took a handful of constructs to build and characterize and shared data to determine which one was working the best to move forward with,” Johnson says.

Nicole Johnson, Ph.D. student in biochemistry, conducts research in June 2021 on campus.
Goldsmith and Hsieh noticed that one combination stood out among the rest for its surprisingly high yield and stability. Their test results indicated that mixing individual proline mutations resulted in particularly stable spikes. The graduate students extensively tested the standout variant and found that the protein boasted excellent scalability and resistance to stressors, thus making it a strong vaccine candidate.
The swift development of HexaPro did not come without setbacks. Broken equipment presented a particularly challenging obstacle for the students, as repair technicians were inaccessible during phases of lockdown. The team also had to improvise to overcome a lack of supplies as the pandemic wreaked havoc on manufacturing and shipping operations worldwide.
Despite the many challenges, the team managed to maintain a cooperative environment. “I’m most proud of my lab for sticking it out during long hours of hard work to solve these problems as fast as possible,” Goldsmith says. “This lab culture, and the amazingly organized systems we have that were set up by Jason and are constantly maintained by all of us, allowed us to excel when the pressure was high. We had to persevere through some very tough times. Still, we always figured out a way to move forward and solve problems.”